scanpy.pl.rank_genes_groups
- scanpy.pl.rank_genes_groups(adata, groups=None, n_genes=20, gene_symbols=None, key='rank_genes_groups', fontsize=8, ncols=4, sharey=True, show=None, save=None, ax=None, **kwds)
Plot ranking of genes.
- Parameters
- adata :
AnnDataAnnData Annotated data matrix.
- groups :
str|Sequence[str] |NoneUnion[str,Sequence[str],None] (default:None) The groups for which to show the gene ranking.
- gene_symbols :
str|NoneOptional[str] (default:None) Key for field in
.varthat stores gene symbols if you do not want to use.var_names.- n_genes :
intint(default:20) Number of genes to show.
- fontsize :
intint(default:8) Fontsize for gene names.
- ncols :
intint(default:4) Number of panels shown per row.
- sharey :
boolbool(default:True) Controls if the y-axis of each panels should be shared. But passing
sharey=False, each panel has its own y-axis range.- show :
bool|NoneOptional[bool] (default:None) Show the plot, do not return axis.
- save :
bool|NoneOptional[bool] (default:None) If
Trueor astr, save the figure. A string is appended to the default filename. Infer the filetype if ending on {'.pdf','.png','.svg'}.- ax :
Axes|NoneOptional[Axes] (default:None) A matplotlib axes object. Only works if plotting a single component.
- adata :
Examples
import scanpy as sc adata = sc.datasets.pbmc68k_reduced() sc.pl.rank_genes_groups(adata)
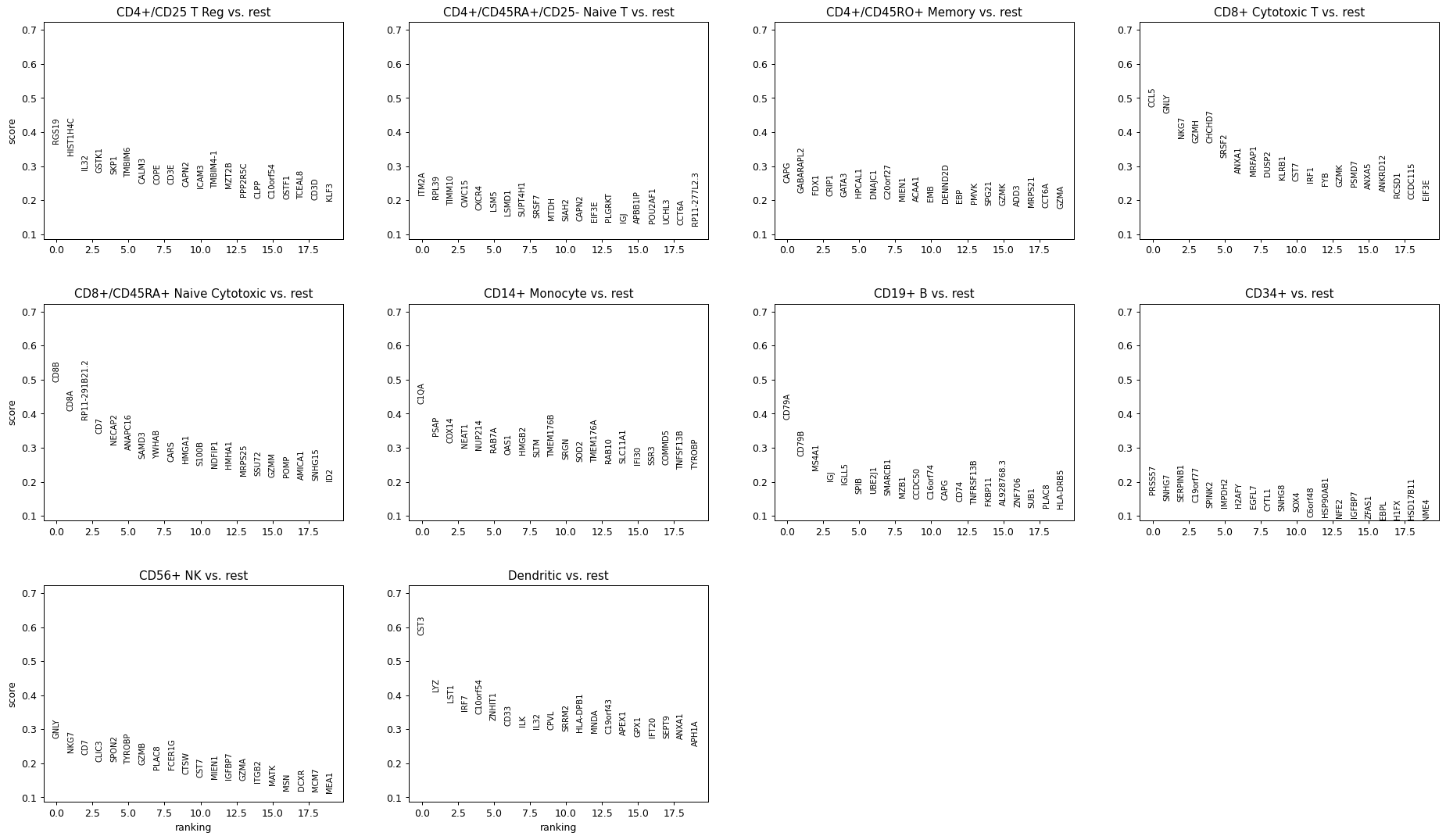
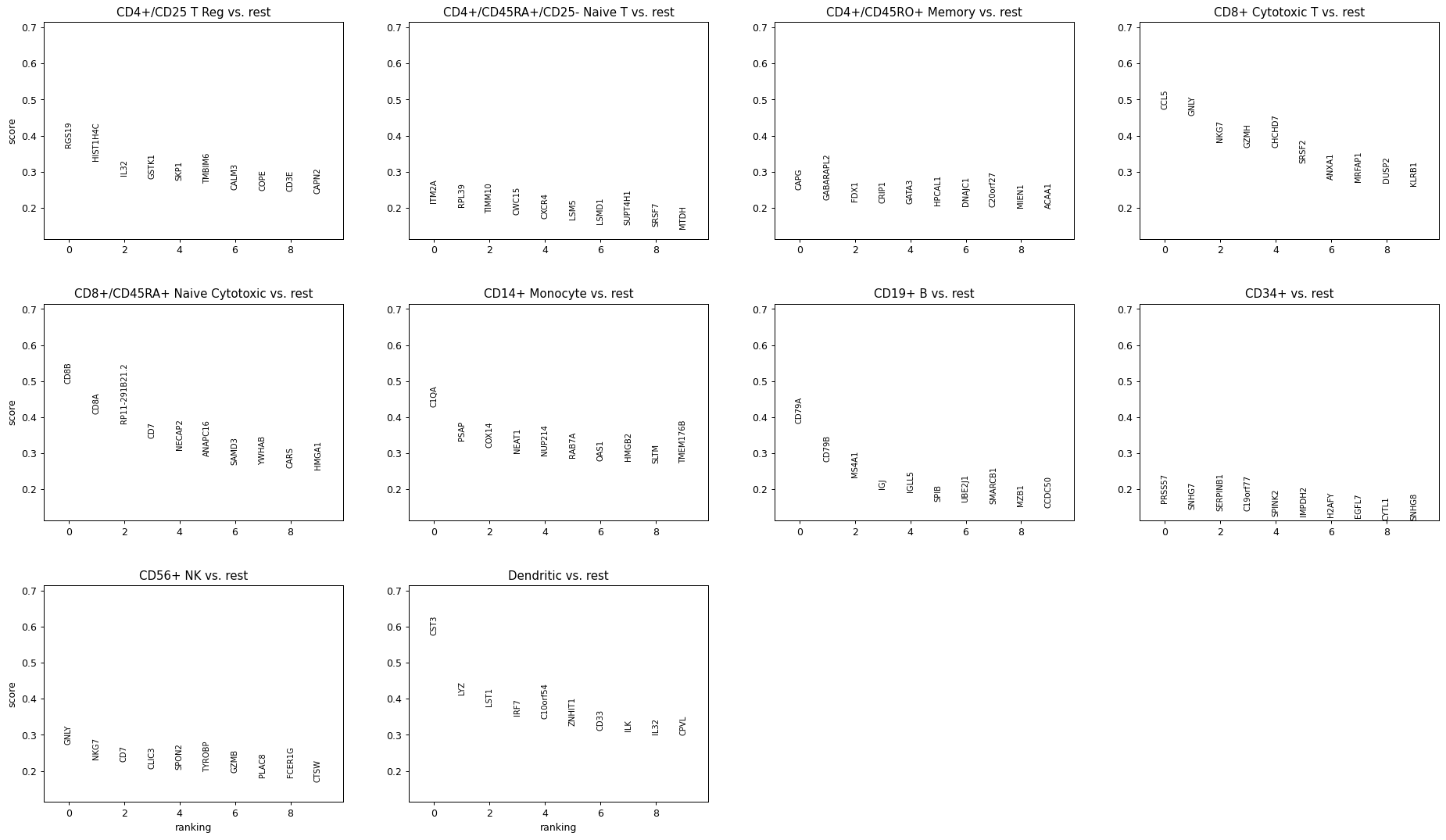
Plot top 10 genes (default 20 genes)
sc.pl.rank_genes_groups(adata, n_genes=10)
See also