scanpy.pl.heatmap#
- scanpy.pl.heatmap(adata, var_names, groupby, *, use_raw=None, log=False, num_categories=7, dendrogram=False, gene_symbols=None, var_group_positions=None, var_group_labels=None, var_group_rotation=None, layer=None, standard_scale=None, swap_axes=False, show_gene_labels=None, show=None, save=None, figsize=None, vmin=None, vmax=None, vcenter=None, norm=None, **kwds)[source]#
Heatmap of the expression values of genes.
If
groupbyis given, the heatmap is ordered by the respective group. For example, a list of marker genes can be plotted, ordered by clustering. If thegroupbyobservation annotation is not categorical the observation annotation is turned into a categorical by binning the data into the number specified innum_categories.- Parameters:
- adata
AnnData Annotated data matrix.
- var_names
str|Sequence[str] |Mapping[str,str|Sequence[str]] var_namesshould be a valid subset ofadata.var_names. Ifvar_namesis a mapping, then the key is used as label to group the values (seevar_group_labels). The mapping values should be sequences of validadata.var_names. In this case either coloring or ‘brackets’ are used for the grouping of var names depending on the plot. Whenvar_namesis a mapping, then thevar_group_labelsandvar_group_positionsare set.- groupby
str|Sequence[str] The key of the observation grouping to consider.
- use_raw
bool|None(default:None) Use
rawattribute ofadataif present.- log
bool(default:False) Plot on logarithmic axis.
- num_categories
int(default:7) Only used if groupby observation is not categorical. This value determines the number of groups into which the groupby observation should be subdivided.
- categories_order
Order in which to show the categories. Note: add_dendrogram or add_totals can change the categories order.
- figsize
tuple[float,float] |None(default:None) Figure size when
multi_panel=True. Otherwise thercParam['figure.figsize]value is used. Format is (width, height)- dendrogram
bool|str(default:False) If True or a valid dendrogram key, a dendrogram based on the hierarchical clustering between the
groupbycategories is added. The dendrogram information is computed usingscanpy.tl.dendrogram(). Iftl.dendrogramhas not been called previously the function is called with default parameters.- gene_symbols
str|None(default:None) Column name in
.varDataFrame that stores gene symbols. By defaultvar_namesrefer to the index column of the.varDataFrame. Setting this option allows alternative names to be used.- var_group_positions
Sequence[tuple[int,int]] |None(default:None) Use this parameter to highlight groups of
var_names. This will draw a ‘bracket’ or a color block between the given start and end positions. If the parametervar_group_labelsis set, the corresponding labels are added on top/left. E.g.var_group_positions=[(4,10)]will add a bracket between the fourthvar_nameand the tenthvar_name. By giving more positions, more brackets/color blocks are drawn.- var_group_labels
Sequence[str] |None(default:None) Labels for each of the
var_group_positionsthat want to be highlighted.- var_group_rotation
float|None(default:None) Label rotation degrees. By default, labels larger than 4 characters are rotated 90 degrees.
- layer
str|None(default:None) Name of the AnnData object layer that wants to be plotted. By default adata.raw.X is plotted. If
use_raw=Falseis set, thenadata.Xis plotted. Iflayeris set to a valid layer name, then the layer is plotted.layertakes precedence overuse_raw.- standard_scale
Optional[Literal['var','obs']] (default:None) Whether or not to standardize that dimension between 0 and 1, meaning for each variable or observation, subtract the minimum and divide each by its maximum.
- swap_axes
bool(default:False) By default, the x axis contains
var_names(e.g. genes) and the y axis thegroupbycategories (if any). By settingswap_axesthen x are thegroupbycategories and y thevar_names.- show_gene_labels
bool|None(default:None) By default gene labels are shown when there are 50 or less genes. Otherwise the labels are removed.
- show
bool|None(default:None) Show the plot, do not return axis.
- save
str|bool|None(default:None) If
Trueor astr, save the figure. A string is appended to the default filename. Infer the filetype if ending on {'.pdf','.png','.svg'}.- ax
A matplotlib axes object. Only works if plotting a single component.
- vmin
float|None(default:None) The value representing the lower limit of the color scale. Values smaller than vmin are plotted with the same color as vmin.
- vmax
float|None(default:None) The value representing the upper limit of the color scale. Values larger than vmax are plotted with the same color as vmax.
- vcenter
float|None(default:None) The value representing the center of the color scale. Useful for diverging colormaps.
- norm
Normalize|None(default:None) Custom color normalization object from matplotlib. See
https://matplotlib.org/stable/tutorials/colors/colormapnorms.htmlfor details.- **kwds
Are passed to
matplotlib.pyplot.imshow().
- adata
- Return type:
- Returns:
Dict of
Axes
Examples
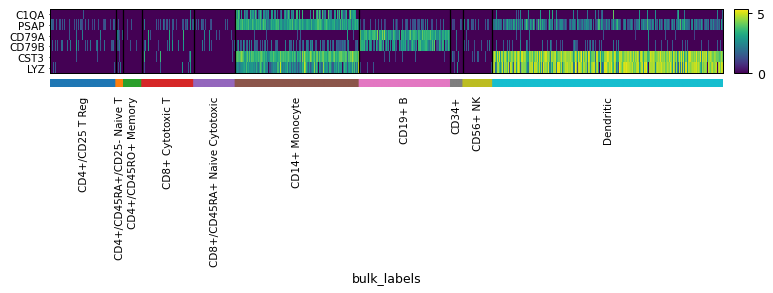
import scanpy as sc adata = sc.datasets.pbmc68k_reduced() markers = ['C1QA', 'PSAP', 'CD79A', 'CD79B', 'CST3', 'LYZ'] sc.pl.heatmap(adata, markers, groupby='bulk_labels', swap_axes=True)