scanpy.pl.pca
- scanpy.pl.pca(adata, *, annotate_var_explained=False, show=None, return_fig=None, save=None, **kwargs)
Scatter plot in PCA coordinates.
Use the parameter
annotate_var_explainedto annotate the explained variance.- Parameters:
- adata
Annotated data matrix.
- color
Keys for annotations of observations/cells or variables/genes, e.g.,
'ann1'or['ann1', 'ann2'].- gene_symbols
Column name in
.varDataFrame that stores gene symbols. By defaultvar_namesrefer to the index column of the.varDataFrame. Setting this option allows alternative names to be used.- use_raw
Use
.rawattribute ofadatafor coloring with gene expression. IfNone, defaults toTrueiflayerisn’t provided andadata.rawis present.- layer
Name of the AnnData object layer that wants to be plotted. By default adata.raw.X is plotted. If
use_raw=Falseis set, thenadata.Xis plotted. Iflayeris set to a valid layer name, then the layer is plotted.layertakes precedence overuse_raw.- annotate_var_explained :
bool(default:False) - sort_order
For continuous annotations used as color parameter, plot data points with higher values on top of others.
- groups
Restrict to a few categories in categorical observation annotation. The default is not to restrict to any groups.
- dimensions
0-indexed dimensions of the embedding to plot as integers. E.g. [(0, 1), (1, 2)]. Unlike
components, this argument is used in the same way ascolors, e.g. is used to specify a single plot at a time. Will eventually replace the components argument.- components
For instance,
['1,2', '2,3']. To plot all available components usecomponents='all'.- projection
Projection of plot (default:
'2d').- legend_loc
Location of legend, either
'on data','right margin'or a valid keyword for thelocparameter ofLegend.- legend_fontsize
Numeric size in pt or string describing the size. See
set_fontsize().- legend_fontweight
Legend font weight. A numeric value in range 0-1000 or a string. Defaults to
'bold'iflegend_loc == 'on data', otherwise to'normal'. Seeset_fontweight().- legend_fontoutline
Line width of the legend font outline in pt. Draws a white outline using the path effect
withStroke.- colorbar_loc
Where to place the colorbar for continous variables. If
None, no colorbar is added.- size
Point size. If
None, is automatically computed as 120000 / n_cells. Can be a sequence containing the size for each cell. The order should be the same as in adata.obs.- color_map
Color map to use for continous variables. Can be a name or a
Colormapinstance (e.g."magma”,"viridis"ormpl.cm.cividis), seeget_cmap(). IfNone, the value ofmpl.rcParams["image.cmap"]is used. The defaultcolor_mapcan be set usingset_figure_params().- palette
Colors to use for plotting categorical annotation groups. The palette can be a valid
ListedColormapname ('Set2','tab20', …), aCyclerobject, a dict mapping categories to colors, or a sequence of colors. Colors must be valid to matplotlib. (seeis_color_like()). IfNone,mpl.rcParams["axes.prop_cycle"]is used unless the categorical variable already has colors stored inadata.uns["{var}_colors"]. If provided, values ofadata.uns["{var}_colors"]will be set.- na_color
Color to use for null or masked values. Can be anything matplotlib accepts as a color. Used for all points if
color=None.- na_in_legend
If there are missing values, whether they get an entry in the legend. Currently only implemented for categorical legends.
- frameon
Draw a frame around the scatter plot. Defaults to value set in
set_figure_params(), defaults toTrue.- title
Provide title for panels either as string or list of strings, e.g.
['title1', 'title2', ...].- vmin
The value representing the lower limit of the color scale. Values smaller than vmin are plotted with the same color as vmin. vmin can be a number, a string, a function or
None. If vmin is a string and has the formatpN, this is interpreted as a vmin=percentile(N). For example vmin=’p1.5’ is interpreted as the 1.5 percentile. If vmin is function, then vmin is interpreted as the return value of the function over the list of values to plot. For example to set vmin tp the mean of the values to plot,def my_vmin(values): return np.mean(values)and then setvmin=my_vmin. If vmin is None (default) an automatic minimum value is used as defined by matplotlibscatterfunction. When making multiple plots, vmin can be a list of values, one for each plot. For examplevmin=[0.1, 'p1', None, my_vmin]- vmax
The value representing the upper limit of the color scale. The format is the same as for
vmin.- vcenter
The value representing the center of the color scale. Useful for diverging colormaps. The format is the same as for
vmin. Example: sc.pl.umap(adata, color=’TREM2’, vcenter=’p50’, cmap=’RdBu_r’)- add_outline
If set to True, this will add a thin border around groups of dots. In some situations this can enhance the aesthetics of the resulting image
- outline_color
Tuple with two valid color names used to adjust the add_outline. The first color is the border color (default: black), while the second color is a gap color between the border color and the scatter dot (default: white).
- outline_width
Tuple with two width numbers used to adjust the outline. The first value is the width of the border color as a fraction of the scatter dot size (default: 0.3). The second value is width of the gap color (default: 0.05).
- ncols
Number of panels per row.
- wspace
Adjust the width of the space between multiple panels.
- hspace
Adjust the height of the space between multiple panels.
- return_fig :
Optional[bool] (default:None) Return the matplotlib figure.
- kwargs
Arguments to pass to
matplotlib.pyplot.scatter(), for instance: the maximum and minimum values (e.g.vmin=-2, vmax=5).- show :
Optional[bool] (default:None) Show the plot, do not return axis.
- save :
Union[str,bool,None] (default:None) If
Trueor astr, save the figure. A string is appended to the default filename. Infer the filetype if ending on {'.pdf','.png','.svg'}.- ax
A matplotlib axes object. Only works if plotting a single component.
- Return type:
- Returns:
: If
show==FalseaAxesor a list of it.
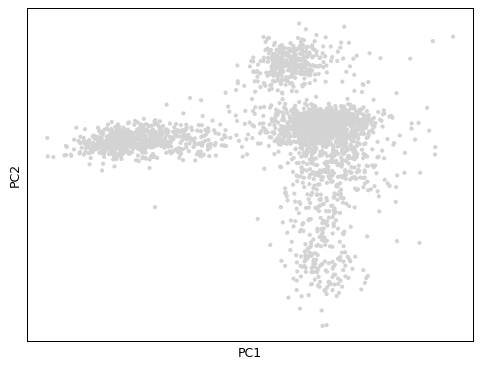
Examples
import scanpy as sc adata = sc.datasets.pbmc3k_processed() sc.pl.pca(adata)
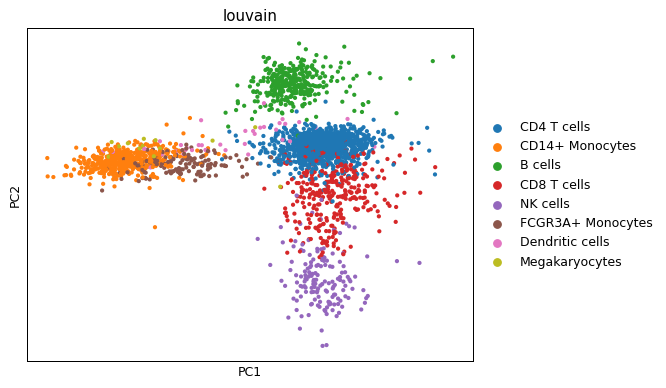
Colour points by discrete variable (Louvain clusters).
sc.pl.pca(adata, color="louvain")
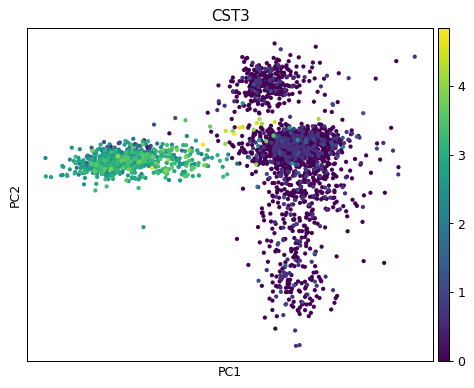
Colour points by gene expression.
sc.pl.pca(adata, color="CST3")